Abstract
Background: Sickle cell disease (SCD) is a devastating Mendelian disease affecting millions of people worldwide. Although SCD is caused by a single-point mutation (Glu6Val) in the β-globin chain of hemoglobin, the clinical manifestations are heterogeneous and systemic, impacting the functions of multiple organs and leading to early mortality. Predicting disease severity and identifying the molecular mechanisms underlying disease pathology and progression remain a challenge.
Objective: The goal of this work was to characterize the clinical heterogeneity associated with SCD using transcriptomics.
Methods: We integrated publicly available bulk and single-cell RNA sequencing (scRNA-seq) data sets that included samples from pediatric to young adult patients and contained clinical phenotypes to identify molecular signatures of SCD severity and heterogeneity (Table). Our integrative transcriptomics analyses included the following:
Identify cell-type clusters and derive novel progenitor signatures from an integrative analysis of the 2 independent scRNA-seq data sets capturing CD34+-enriched or unselected bone marrow (BM) cells from SCD and control samples.
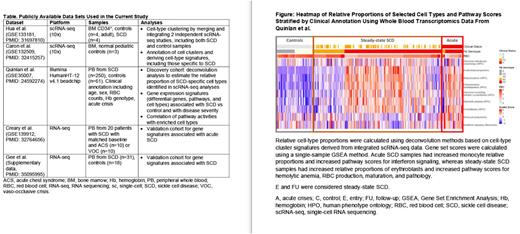
Calculate the relative proportions of cell types in whole blood by deconvolution using both the scRNA-seq-derived novel cell-type signatures (dtangle) and publicly available immune cell-type signatures (xCell, EPIC, quanTIseq).
Identify signatures (genes, pathways, cell types) associated with disease severity and hemoglobin genotypes using various bioinformatics approaches, including differential gene expression and single-sample Gene Set Enrichment Analysis, among others, and examine relationships of enriched cell types and pathways (queried from MSigDB) in discovery followed by confirmation in validation data sets.
Results: Cell-type clusters and gene signatures were generated by combining and integrating scRNA-seq data sets from BM SCD and control samples (Table). Using our fine-mapped cell-type clusters, we found an erythroid progenitor subclass to be depleted, whereas a lymphoid progenitor subclass expanded in SCD BM samples, increasing granularity of the cell types reported by Hua et al.
Cell-type deconvolution of bulk transcriptome data using the combined scRNA-seq-derived signatures revealed enriched monocyte relative proportions in acute crisis compared with steady-state SCD samples (β=0.01, P=1.13×10−7) and enriched erythroblast relative proportions in SCD compared with control samples (β=0.02, P=1.10×10−12). Deconvolution using publicly available immune cell signatures showed significant enrichment of neutrophil and macrophage signatures in acute SCD samples (Padj<0.1).
Analysis of bulk RNA-seq data indicated a set of highly correlated genes differentially expressed in steady-state SCD and associated with varied pathways, including red blood cell (RBC) morphology, hematopoiesis, anemia, and cholelithiasis. Higher pathway scores were seen in severe (HbSS) genotype samples than in milder (HbSC) genotype samples, consistent with known disease pathology and severity (Figure). Building on results from integrative analyses, we found that interferon and inflammatory response genes were enriched in acute SCD in both discovery and validation data sets. We found significant correlations between monocyte relative proportions and interferon signaling pathway scores, and between erythroblast relative proportions and the pathway scores affecting RBC production, morphology, and pathology and potential SCD comorbidities such as cholelithiasis (Figure).
Conclusions: Our integrative scRNA-seq analysis led to robust cell cluster identification to improve deconvolution analysis of bulk transcriptomics data. We found elevated interferon and inflammatory pathway activity in acute SCD samples, consistent with findings in the literature (PMIDs: 34166491, 32764656). Furthermore, we found enriched neutrophils and monocyte signatures in acute SCD, suggesting their role in mediating the innate immune response. These findings provide motivation to characterize the role of neutrophils and monocytes in acute SCD crises. Our integrative transcriptomics analyses showed defective RBC production and maturation in SCD. Using the signatures from this work, we aim to elucidate the heterogeneity within SCD pathophysiology.
Funding: Global Blood Therapeutics.
Disclosures
Potdar:Global Blood Therapeutics: Current Employment. Caponegro:Vindhya Data Science: Current Employment. Soylemez:Global Blood Therapeutics: Current Employment. Reddy:Vindhya Data Science: Current Employment. Yu:Global Blood Therapeutics: Current Employment.
Author notes
Asterisk with author names denotes non-ASH members.